The KEGG (Kyoto Encyclopedia of Genes and Genomes) database is a useful reference of biochemical pathways often employed in metabolomics and proteomics studies. Various tables from the database can be downloaded in flat file format via ftp (ftp://ftp.pathway.jp/kegg). In addition, xml files containing information on pathway reactions can be download. Although these provide a useful interface to query properties, the real interest in pathways is in their connectivity.
Of the KEGG tables, the following are most of interest:
- reaction: Information on individual reactions (id tag(entry), reaction name, definition of reaction, equation, list of rclass, pathways on which it can be found, enzyme involved in reaction)
- rclass: Information on different reaction classes (id tag (entry), definition of reaction class, pathways on which it can be found, series of pairs of compounds which undergo the reaction (substrate and product pairs), reactions in which this reaction class can be found)
- compound: Information on compounds (id tag (entry), name, formula, accurate mass, pathways on which it can be found)
- enzyme: Information on enzymes (id tag (entry), name, reaction in which its involved, substrate and product information, pathways on which it can be found)
The xml files contain three types of element, namely entry (compounds and enzymes), relation (connections between entries) and *reaction^ (information on reactions between substrates/products).
There’s considerable redundancy between the tables and it’s relatively straightforward to create a neo4j database from these pieces of information.
The command used to populate the database is simply:
"MERGE (c1:Compound{properties}) MERGE (c2:Compound{properties}) CREATE (c1)-[:REACTION{properties}]->(c2)"
where properties refer to the properties associated with nodes and relations (in JSON notation).
To run this within a transaction it’s:
tx = graph.begin()
for t in triples:
mergeText = "MERGE (c1:Compound{properties}) MERGE (c2:Compound{properties}) CREATE (c1)-[:REACTION{properties}]->(c2)"
tx.append(mergeText)
tx.commit()
replacing the properies with properties of each reaction.
Three neo4j databases were constructed, comparing the success and information available in each case.
construct from rclass file
The most straightforward approach is to use the data stored in the rclass file. Each rclass record contains an identifier (entry) and pairs of molecules which undergo the reaction. The database is constructed by Simply taking the pairs of molecules and constructing a series of merge and create records. This method is successful, however the records do not contain information about the direction of the reaction (A->B, A<-B, A<->B).
construct from xml files
The xml files contain data on the nodes (compounds / enzymes) and relationships (reactions). They also include the reaction direction (reversible or irreversible). Once read and parsed, the reaction information can be used to populate the database (cross-referencing to information in additional tables). Upon inspection, however, its apparent that the xml files are missing some connectivity information. Indeed, the files have been constructed to aid in visual interpretation of the data which leads to redundancy and missing information.
| Mapping Connections | Mapping Reactions |
|---|---|
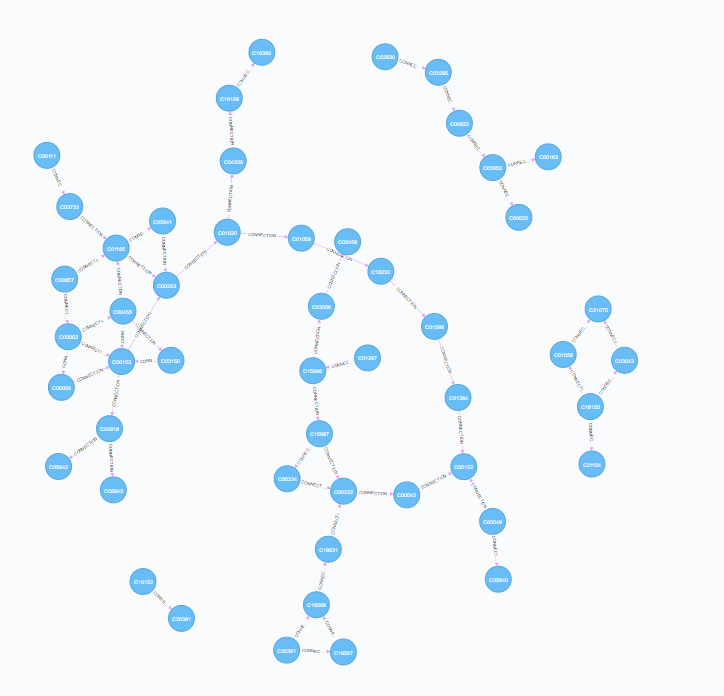 |
 |
construct from reaction and file
Cross-referencing the reaction file with the compound, rclass and enzyme files provides the richest information. Two types of relationships can be constructed here - each individual reaction (multiple connections between each pair of compounds) as well as single points of connection between each pair (equivalent to rclass). This highlights the ability of neo4j to store multiple types of relationship between nodes.
| Mapping Connections | Mapping Reactions |
|---|---|
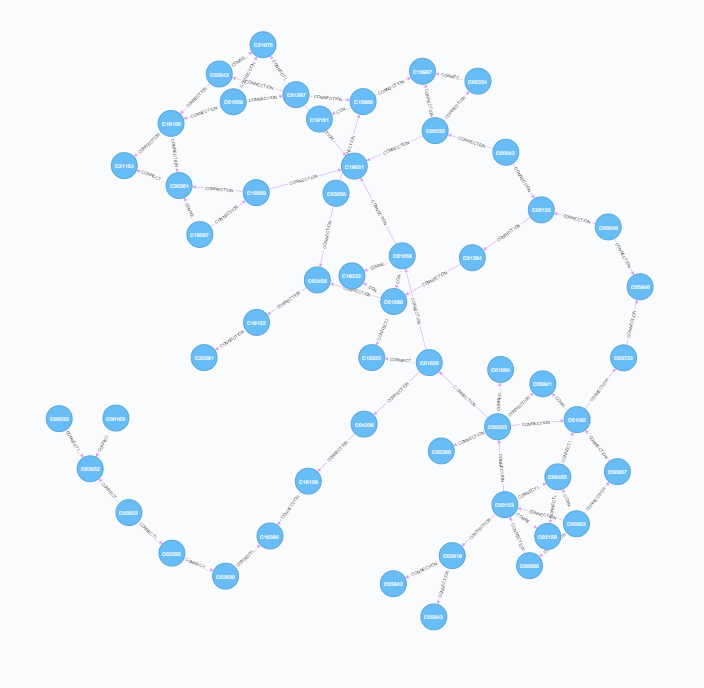 |
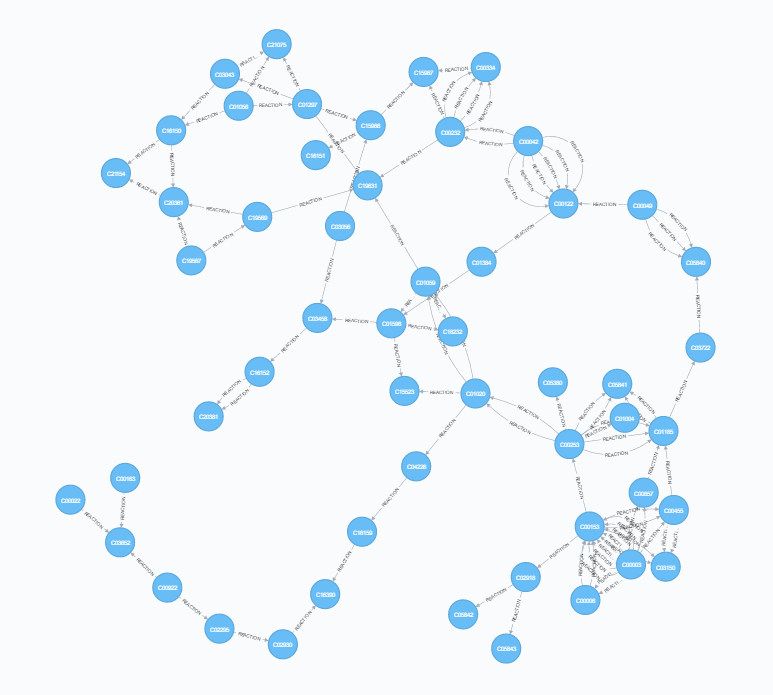 |
The code can be found at https://github.com/harveyl888/neo4jKEGG